Multistart nonlinear least squares fitting with {gslnls}
Introduction
When fitting a nonlinear regression model in R with nls(), the first step is to select an appropriate regression model to fit the observed data, the second step is to find reasonable starting values for the model parameters in order to initialize the nonlinear least-squares (NLS) algorithm. In some cases, choosing starting values is straightforward, for instance when there is some physical interpretation to the model or the least squares objective function is highly regular and easy to optimize. In other cases, this can be more challenging, especially when the least squares objective consists of large plateaus with local minima located in small narrow canyons (see e.g. Transtrum, Machta, and Sethna (2011)), or when the parameters live on very different scales. To illustrate, consider the following Hobbs’ weed infestation example dataset from (Nash 1979) and (Nash 2014):
## Hobbs' weed infestation data
hobbs_weed <- data.frame(
x = 1:12,
y = c(5.308, 7.24, 9.638, 12.866, 17.069, 23.192, 31.443, 38.558, 50.156, 62.948, 75.995, 91.972)
)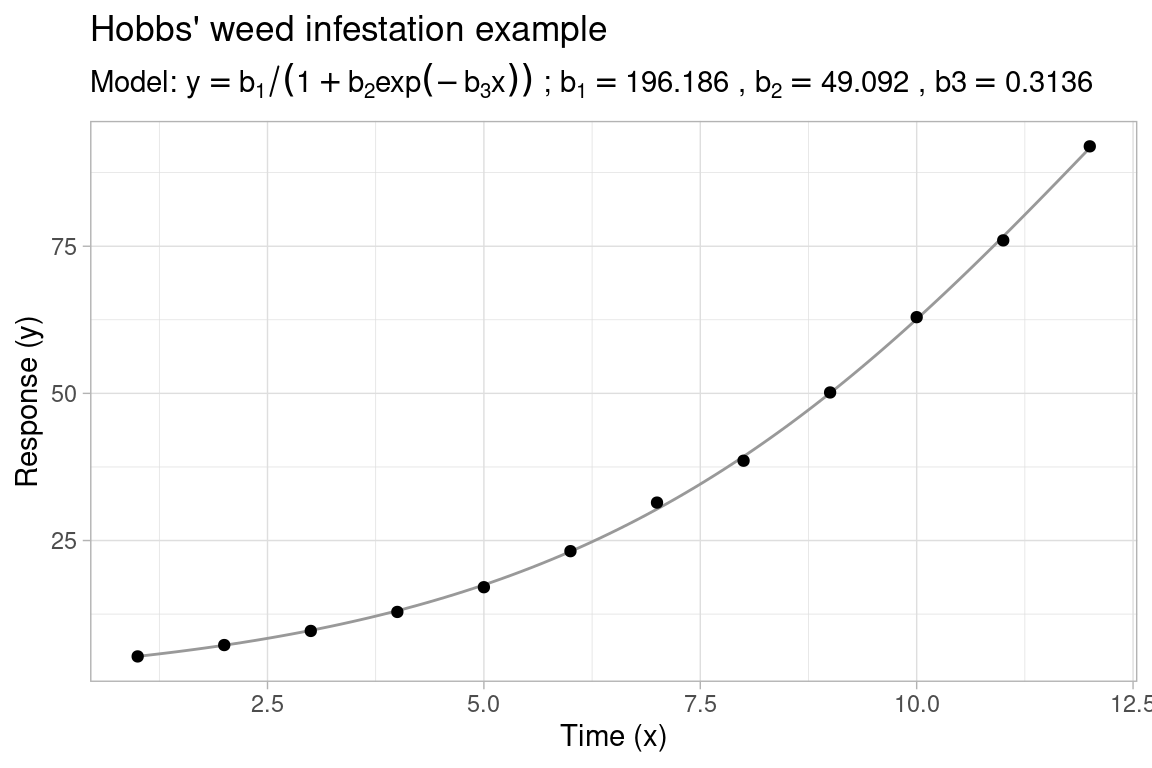
Using the standard Gauss-Newton algorithm to fit the nonlinear model y ~ b1 / (1 + b2 * exp(-b3 * x)) with a basic nls() call, correct convergence of the algorithm is (perhaps) surprisingly sensitive to the choice of starting values. If not properly initialized, the parameters tend to run away to infinity, also known as parameter evaporation:
## initial attempt nls
nls(
formula = y ~ b1 / (1 + b2 * exp(-b3 * x)),
data = hobbs_weed,
start = c(b1 = 100, b2 = 10, b3 = 1),
trace = TRUE
)
#> 34094.85 (3.11e+00): par = (100 10 1)
#> 11186.49 (3.86e+00): par = (78.18911 3.290385 0.4759647)
#> 5112.607 (4.69e+01): par = (80.9356 3.413601 0.1187296)
#> 5032.864 (3.53e+01): par = (114.9669 5.478666 0.1037978)
#> 4937.315 (2.78e+01): par = (184.1488 9.704171 0.09525315)
#> 4798.508 (2.41e+01): par = (346.9847 19.72996 0.09116909)
#> 4624.151 (2.16e+01): par = (1325.61 80.41635 0.08932604)
#> 3859.425 (1.91e+01): par = (23585.2 1465.277 0.09589796)
#> 1605.353 (1.32e+01): par = (7709100 479913.4 0.1272658)
#> Error in nls(formula = y ~ b1/(1 + b2 * exp(-b3 * x)), data = hobbs_weed, : singular gradientThis is in fact an example where the Gauss-Newton direction initially points to the boundary of the parameter space (see also Figure 7 in Transtrum, Machta, and Sethna (2011)). Instead, using a damped least squares algorithm (Levenberg-Marquardt) that combines both the Gauss-Newton and gradient descent directions, we expect the parameters to evaporate less quickly:
library(gslnls)
## initial attempt gsl_nls
gsl_nls(
fn = y ~ b1 / (1 + b2 * exp(-b3 * x)),
data = hobbs_weed,
start = c(b1 = 100, b2 = 10, b3 = 1),
algorithm = "lm"
)
#> Nonlinear regression model
#> model: y ~ b1/(1 + b2 * exp(-b3 * x))
#> data: hobbs_weed
#> b1 b2 b3
#> 196.1863 49.0916 0.3136
#> residual sum-of-squares: 2.587
#>
#> Algorithm: multifit/levenberg-marquardt, (scaling: more, solver: qr)
#>
#> Number of iterations to convergence: 15
#> Achieved convergence tolerance: 1.554e-14In an attempt to reduce the dependence of the NLS algorithm on a single set of (poorly selected) starting values, this post demonstrates a new multistart procedure available1 in R-package gslnls, which can be useful when we only have limited knowledge regarding the expected parameter values or when we wish to automate nonlinear model fits across multiple different datasets.
A common approach in R to avoid the need for user-supplied NLS starting values is to make use of so-called selfStart models (SSasymp(), SSfpl(), SSmicmen(), etc.) that include an initialization function to return reasonable starting values for the nonlinear model given the available data. The initialization function typically considers a simpler approximate or linearized version of the nonlinear model for which parameters can be estimated without starting values, using e.g. lm(). This is the model fitting approach that is utilized by drc and nlraa among other packages. If we intend to fit a nonlinear model for which a selfStart implementation is already available (or straightforward to implement), this is definitely the recommended approach, since the starting values obtained from a selfStart model are usually well-informed and most NLS routines will not have trouble obtaining the correct parameter estimates from these starting values.
If no selfStart model is available, another approach is to repeatedly call nls() using different starting values drawn as random or fixed points from a pre-defined grid. This is implemented by nls2::nls2() using one of the methods "brute-force", "grid-search", or "random-search". The new multistart procedure in gsl_nls() tries to improve on naive random or grid-based multistart optimization, which can be a very time consuming process, especially if the number of parameters is large (curse of dimensionality) or the scale of the parameter ranges to evaluate is quite broad. As in any multistart global optimization procedure, the ideal approach would be to start a local NLS optimizer within each basin of attraction2 exactly once to reach all existing local minima with minimal computational effort. In practice, we try to avoid running too many local optimizers in the same basin of attraction (resulting in the same local minimum) by exploring the parameter space for promising starting values that might converge to an unseen (local) optimum. The multistart algorithm implemented in gsl_nls() is a modified version of the algorithm in (Hickernell and Yuan 1997) that works both with or without a pre-defined grid of starting ranges. Before describing the details of the multistart procedure, below are several examples illustrating its usage with gsl_nls().
NLS examples
Hobbs’ weed infestation example
Revisiting the Hobbs’ weed infestation example above, we fit the nonlinear model with gsl_nls() using a (fixed) set of starting ranges for the parameters instead of individual starting values:
## multistart attempt gsl_nls
gsl_nls(
fn = y ~ b1 / (1 + b2 * exp(-b3 * x)),
data = hobbs_weed,
start = list(b1 = c(0, 1000), b2 = c(0, 1000), b3 = c(0, 10))
)
#> Nonlinear regression model
#> model: y ~ b1/(1 + b2 * exp(-b3 * x))
#> data: hobbs_weed
#> b1 b2 b3
#> 196.1863 49.0916 0.3136
#> residual sum-of-squares: 2.587
#>
#> Algorithm: multifit/levenberg-marquardt, (scaling: more, solver: qr)
#>
#> Number of iterations to convergence: 3
#> Achieved convergence tolerance: 2.842e-14Alternatively, we can leave one or more starting ranges undefined, in which case they are updated dynamically during the multistart optimization:
## multistart attempt 2 gsl_nls
gsl_nls(
fn = y ~ b1 / (1 + b2 * exp(-b3 * x)),
data = hobbs_weed,
start = list(b1 = NA, b2 = NA, b3 = NA)
)
#> Nonlinear regression model
#> model: y ~ b1/(1 + b2 * exp(-b3 * x))
#> data: hobbs_weed
#> b1 b2 b3
#> 196.1863 49.0916 0.3136
#> residual sum-of-squares: 2.587
#>
#> Algorithm: multifit/levenberg-marquardt, (scaling: more, solver: qr)
#>
#> Number of iterations to convergence: 4
#> Achieved convergence tolerance: 5.329e-15NIST StRD Gauss1 example
Second, we consider the Gauss1 regression test problem as listed in the NIST StRD Nonlinear Regression archive. The observed data takes the shape of a camel’s back consisting of two Gaussians on a decaying exponential baseline subject to additive Gaussian noise. The data, nonlinear model and target parameter values are included in the gslnls package and available through the function nls_test_problem():
gauss1 <- nls_test_problem("Gauss1")
## data
str(gauss1$data)
#> 'data.frame': 250 obs. of 2 variables:
#> $ y: num 97.6 97.8 96.6 92.6 91.2 ...
#> $ x: num 1 2 3 4 5 6 7 8 9 10 ...
## model + target parameters
gauss1[c("fn", "target")]
#> $fn
#> y ~ b1 * exp(-b2 * x) + b3 * exp(-(x - b4)^2/b5^2) + b6 * exp(-(x -
#> b7)^2/b8^2)
#> <environment: 0x562f10e8ceb8>
#>
#> $target
#> b1 b2 b3 b4 b5 b6
#> 98.77821087 0.01049728 100.48990633 67.48111128 23.12977336 71.99450300
#> b7 b8
#> 178.99805021 18.38938902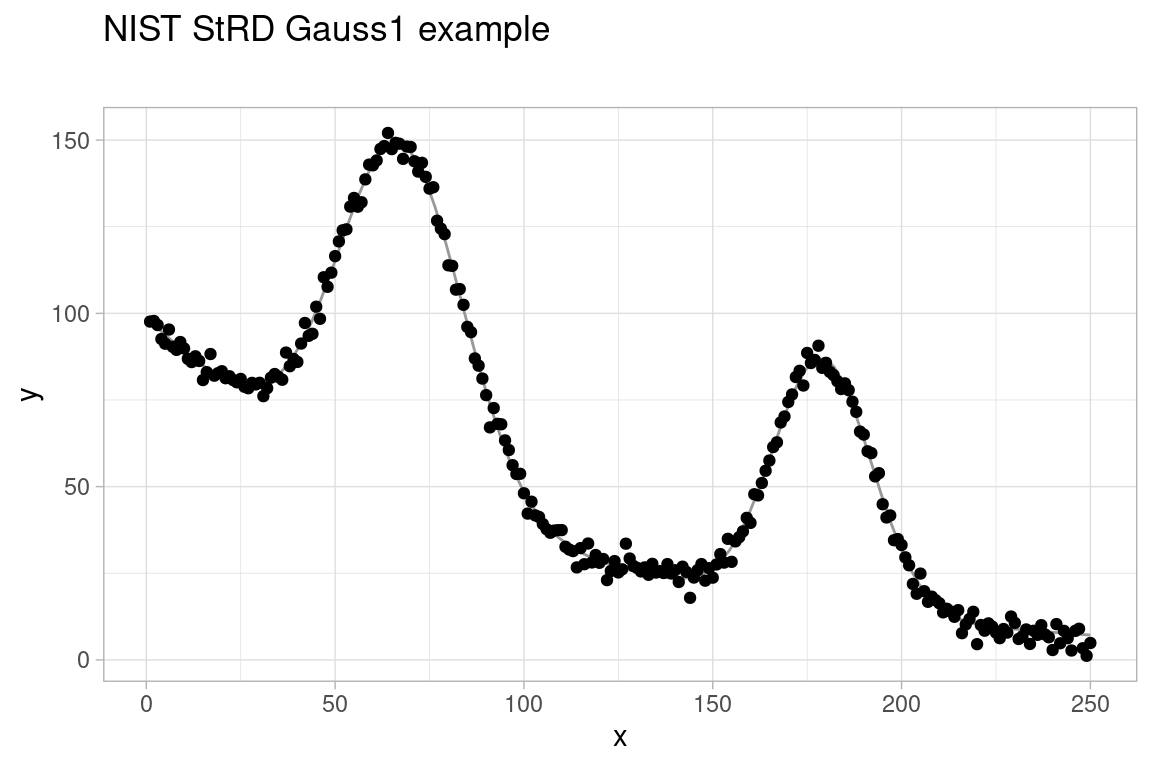
Due to the large number of parameters in the model y ~ b1 * exp(-b2 * x) + b3 * exp(-(x - b4)**2 / b5**2) + b6 * exp(-(x - b7)**2 / b8**2), finding starting values from which nls() is able to correctly fit the model is a tedious task. Below is an initial attempt at solving the NLS problem with the NL2SOL algorithm (algorithm = "port") and all parameters bounded from below by zero:
## initial attempt nls
nls(
formula = gauss1$fn,
data = gauss1$data,
start = c(b1 = 100, b2 = 0, b3 = 100, b4 = 75, b5 = 25, b6 = 75, b7 = 125, b8 = 25),
lower = 0,
algorithm = "port"
)
#> Error in nls(formula = gauss1$fn, data = gauss1$data, start = c(b1 = 100, : Convergence failure: singular convergence (7)As seen from this example, most parameter starting values –except for b7– are not that far from their target values. However, nls() still fails to converge due to a single poorly selected starting value for the parameter b7.
Using the multistart procedure in gsl_nls(), we can combine fixed starting values or ranges when good initial guesses for the parameters are available together with missing starting values for the parameters that are lacking such information. This avoids the need to select poor starting values for certain parameters, which may cause the NLS optimization to fail as in our previous attempt to fit the model.
## multistart attempt gsl_nls
gsl_nls(
fn = gauss1$fn,
data = gauss1$data,
start = list(b1 = 100, b2 = c(0, 1), b3 = NA, b4 = NA, b5 = NA, b6 = NA, b7 = NA, b8 = NA),
lower = 0
)
#> Nonlinear regression model
#> model: y ~ b1 * exp(-b2 * x) + b3 * exp(-(x - b4)^2/b5^2) + b6 * exp(-(x - b7)^2/b8^2)
#> data: gauss1$data
#> b1 b2 b3 b4 b5 b6 b7 b8
#> 98.7782 0.0105 100.4899 67.4811 23.1298 71.9945 178.9981 18.3894
#> residual sum-of-squares: 1316
#>
#> Algorithm: multifit/levenberg-marquardt, (scaling: more, solver: qr)
#>
#> Number of iterations to convergence: 4
#> Achieved convergence tolerance: 2.274e-13Lubricant dataset (Bates & Watts)
As a final example, consider the Lubricant dataset from (Bates and Watts 1988, Appendix 1, A1.8), which measures the kinematic viscosity of a lubricant as a function of temperature (°C) and pressure (atm). The Lubricant data, model and target parameter are included as an NLS test problem in gslnls and can be retrieved with nls_test_problem() similar to the previous example. Here, x1 encodes the temperature predictor (in °C) and x2 is the pressure (atm) predictor.
lubricant <- nls_test_problem("Lubricant")
## data
str(lubricant$data)
#> 'data.frame': 53 obs. of 3 variables:
#> $ y : num 5.11 6.39 7.39 5.79 5.11 ...
#> $ x1: num 0 0 0 0 0 0 0 0 0 0 ...
#> $ x2: num 0.001 0.741 1.407 0.363 0.001 ...
## model + target parameters
lubricant[c("fn", "target")]
#> $fn
#> y ~ b1/(b2 + x1) + b3 * x2 + b4 * x2^2 + b5 * x2^3 + (b6 * x2 +
#> b7 * x2^3) * exp(-x1/(b8 + b9 * x2^2))
#> <environment: 0x562f0ab01c30>
#>
#> $target
#> b1 b2 b3 b4 b5
#> 1054.54053186 206.54577890 1.46031479 -0.25965483 0.02257371
#> b6 b7 b8 b9
#> 0.40138428 0.03528405 57.40463143 -0.47672110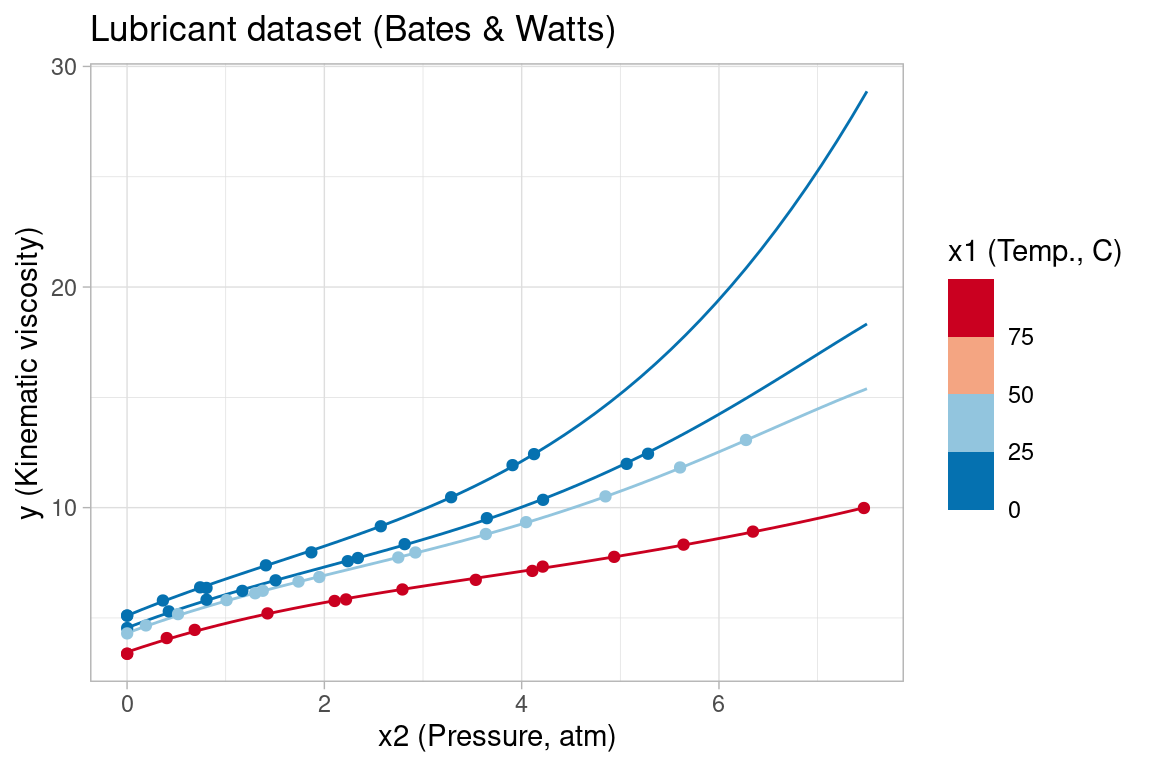
The nonlinear model contains a relatively large number of parameters, similar to the Gauss1 example, and the model fitting is further complicated by the large differences in magnitude of the target parameters. Without a more systematic approach, e.g. by linearizing parts of the model to initialize parameters as in (Bates and Watts 1988, Ch 3.6), it is difficult to obtain a good model fit with nls() by naively trying different sets of starting values. Here, we again use the NL2SOL algorithm (algorithm = "port"), selecting starting values that are all close to the target parameters, but which still cause nls() to fail.
## initial attempt nls
nls(
formula = lubricant$fn,
data = lubricant$data,
start = c(b1 = 1000, b2 = 200, b3 = 1, b4 = -0.01, b5 = 0.01, b6 = 0.01, b7 = 0.01, b8 = 50, b9 = -0.01),
algorithm = "port"
)
#> Error in nls(formula = lubricant$fn, data = lubricant$data, start = c(b1 = 1000, : Convergence failure: false convergence (8)Attempting to solve the NLS problem with the multistart procedure in gsl_nls(), we are able to obtain correct NLS convergence without any knowledge of the target parameters by leaving all starting values unspecified:
## multistart attempt gsl_nls
gsl_nls(
fn = lubricant$fn,
data = lubricant$data,
start = c(b1 = NA, b2 = NA, b3 = NA, b4 = NA, b5 = NA, b6 = NA, b7 = NA, b8 = NA, b9 = NA)
)
#> Nonlinear regression model
#> model: y ~ b1/(b2 + x1) + b3 * x2 + b4 * x2^2 + b5 * x2^3 + (b6 * x2 + b7 * x2^3) * exp(-x1/(b8 + b9 * x2^2))
#> data: lubricant$data
#> b1 b2 b3 b4 b5 b6 b7
#> 1054.54080 206.54584 1.46031 -0.25965 0.02257 0.40138 0.03528
#> b8 b9
#> 57.40467 -0.47672
#> residual sum-of-squares: 0.08702
#>
#> Algorithm: multifit/levenberg-marquardt, (scaling: more, solver: qr)
#>
#> Number of iterations to convergence: 6
#> Achieved convergence tolerance: 5.69e-16Multistart algorithm details
Before evaluating more NLS test problems, this section provides a more comprehensive overview of the implemented multistart algorithm. The implementation is primarily based on the global optimization algorithm in (Hickernell and Yuan 1997), but slightly modified to the context of trust-region nonlinear least squares. The multistart algorithm consists of multiple major iterations. At the start of major iteration \(M\), a set of pseudo-random starting points is sampled inside the grid of starting ranges. Starting points with an almost singular (approximate) Hessian matrix are discarded immediately, as these are unlikely to converge to a local optimum. For the remaining points, a few iterations of the NLS optimizer are applied in order to distinguish promising from unpromising starting points. At each major iteration, only the \(q\) most promising starting points are kept, and if a starting point is not discarded from this set for at least \(s\) major iterations, a full NLS optimization routine is executed using this starting point. If a new optimal solution is found, the number of optimal stationary points NOSP is incremented and the number of found worse stationary points NWSP is reset to zero. If the obtained solution has already been observed before, or if it converges to a local minimum that is worse than the current optimal solution, the number of worse stationary points NSWP is incremented. If the number of found worse stationary points (NWSP) becomes much larger than the number of optimal starting points (NOSP), then it is likely that we have exhausted the search space and are unable to further improve the current optimal solution. The following pseudo-code provides a high-level description of the multistart procedure. For simplicity, we first consider the scenario in which all \(p\) parameter starting ranges \([l_1, u_1] \times \ldots \times [l_{p}, u_{p}]\) are pre-defined and fixed.
- INITIALIZE
- Choose parameters \(N \geq q \geq 1\), \(L \geq \ell \geq 1\), \(s \geq 1\), \(r > 1\),
maxstart\(\geq 1\),minsp\(\geq 1\). Set \(M = 0\), NOSP = NWSP = 0, and initialize a scaling vector \(\boldsymbol{D} = (0.75, \dots, 0.75) \in [0, 1]^p\).
- Choose parameters \(N \geq q \geq 1\), \(L \geq \ell \geq 1\), \(s \geq 1\), \(r > 1\),
- SAMPLE
- Sample \(N\) initial \(p\)-dimensional points \(\boldsymbol{\theta}_1,\ldots, \boldsymbol{\theta}_N\) inside the grid of starting ranges \([l_1, u_1] \times \ldots \times [l_{p}, u_{p}]\) from a pseudo-random Sobol sequence.
- For \(i = 1,\ldots,N\) and \(k = 1,\ldots,p\), rescale the sampled points inside the grid of starting ranges favoring points near zero according to3: \(\theta^*_{i,k} = \frac{\text{sgn}(\theta_{i,k})}{D_k} \cdot ((|\theta_{i,k}| - l_k^+ - u_k^- + 1)^{D_k} - 1) + l_k^+ - u_k^-\).
- REDUCE 1 AND CONCENTRATE
- Discard all points for which \(\det\left(J(\boldsymbol{\theta}^*_i)^TJ(\boldsymbol{\theta}^*_i)\right) < \epsilon\), with \(\epsilon > 0\) small and \(J(\boldsymbol{\theta}^*_i)\) the Jacobian evaluated at \(\boldsymbol{\theta}^*_i\).
- For each remaining point apply (a small number) \(\ell\) iterations of the NLS optimizer, obtaining concentrated points \(\boldsymbol{\theta}_1^{**},\ldots,\boldsymbol{\theta}_{N_1}^{**}\)
- REDUCE 2
- Identify the first \(q\) order statistics of \(F(\boldsymbol{\theta}_1^{**}),\ldots,F(\boldsymbol{\theta}_{N_1}^{**})\), with \(F(\boldsymbol{\theta}_i^{**})\) the NLS objective evaluated at \(\boldsymbol{\theta}_i^{**}\). Denote \(I_q\) for the set of indices (of size \(q\)).
- Update counter \(S(i) = S(i) + 1\) if \(i \in I_q\), otherwise set \(S(i) = 0\).
- OPTIMIZE
- For each \(i\) with \(S(i) \geq s\), if \(F(\boldsymbol{\theta}_i^{**}) < (1 + \delta) \cdot F(\boldsymbol{\theta}^{opt})\), apply \(L\) iterations of the NLS optimizer, obtaining \(\boldsymbol{\theta}_i^{***}\), and set \(S(i) = 0\).
- If \(F(\boldsymbol{\theta}_i^{***}) < F(\boldsymbol{\theta}^{opt})\), set \(\boldsymbol{\theta}^{opt} = \boldsymbol{\theta}_i^{***}\), and update the scaling vector \(\boldsymbol{D}\) by \(D_k = \left(\frac{\min_\kappa \Delta_\kappa}{\Delta_k}\right)^\alpha\), with exponent \(\alpha \in [0, 1]\) and \(\boldsymbol{\Delta} = \text{diag}(J(\boldsymbol{\theta}^{***}_i)^TJ(\boldsymbol{\theta}^{***}_i))\), i.e. the diagonal damping matrix as used by Marquardt’s scaling method. Update the number of found optimal stationary points NOSP = NOSP + 1, and reset the number of found worse stationary points NWSP = 0.
- If \(F(\boldsymbol{\theta}_i^{***}) \geq F(\boldsymbol{\theta}^{opt})\), set NWSP = NWSP + 1.
- REPEAT
- If NWSP \(\geq\) \(r \cdot\) NSP and NSP \(\geq\)
minsp(minimum required number of stationary points) then stop with success. Otherwise, if \(M <\)maxstart, increment \(M = M + 1\) and return to step 2. sampling new pseudo-random points for each \(i\) with \(S(i) = 0\), reusing \(\boldsymbol{\theta}_i = \boldsymbol{\theta}_i^{**}\) if \(S(i) > 0\).
- If NWSP \(\geq\) \(r \cdot\) NSP and NSP \(\geq\)
It is important to point out that there is no guarantee that the NLS objective \(F(\boldsymbol{\theta}^{opt})\) at the solution returned by the multistart procedure indeed evaluates to the global minimum inside the grid of starting ranges. This may for instance be due to the rescaling function in step 2., which is a type of inverse logistic function scaled to the starting range \([l_k, u_k]\). The scaling exponent \(D_k\) is calculated from the damping matrix in Marquardt’s scaling method, thereby rescaling each parameter differently based on an approximate measure of its order of magnitude. If the damping matrix that is used to calculate the \(D_k\)’s is highly –but incorrectly– confident about the order of magnitudes of certain parameters, we may not explore the parameter starting ranges \([l_k, u_k]\) sufficiently broadly in the subsequent major iterations. Increasing the number of sampled points \(N\) at the start of each major iteration may help to overcome such issues. If we suspect that the returned optimal solution \(\boldsymbol{\theta}^{opt}\) is only a local optimizing solution, we can always force the multistart procedure to continue searching until a better optimum is found by increasing minsp (minimum required number of stationary points).
Missing starting ranges
If no fixed starting values or ranges are defined for certain parameters, as demonstrated in the above examples, then the missing ranges \([l_k, u_k]\) are initialized to the unit interval and dynamically increased or decreased in each major iteration of the multistart algorithm. The decision to increase or decrease the limits of a parameter’s starting range is driven by the minimum and maximum parameter values obtained from the \(q\) best-performing concentrated points (step 3. and 4.) with indices included in \(I_q\). These typically provide a rough indication of the order of magnitude of the parameter range in which to search for the optimal solution. If the dynamic parameter ranges fail to grow sufficiently large to include the global optimizing solution, it may help to increase the values of \(N\), \(r\), maxstart or minsp to avoid early termination of the algorithm at the cost of increased computation effort.
NLS test problems
At the moment of writing this post, 59 NLS test problems are included in gslnls originating primarily from the NIST StRD Nonlinear Regression archive, (Bates and Watts 1988) and (Moré, Garbow, and Hillstrom 1981). This collection of test problems contains 33 regression problems, with nonlinear models defined as a formula and the number of parameters and observations fixed (p, n fixed). The other 26 problems are NLS optimization problems, ported from the Fortran library TEST_NLS. For these problems the nonlinear models are defined as a function and some of the models allow for the number of parameters and observations to be freely varied, only requiring that the number of parameters does not exceed the number of observations/residuals (p <= n free and p == n free). The table below lists all 59 test problems as returned by nls_test_list() including their default number of observations and parameters as set in gslnls.
| Dataset name | Reference | # Observations (n) | # Parameters (p) | Data constraint | Model expression |
|---|---|---|---|---|---|
| Misra1a | NIST StRD (1978) | 14 | 2 | p, n fixed | formula |
| Chwirut2 | NIST StRD (1978) | 54 | 3 | p, n fixed | formula |
| Chwirut1 | NIST StRD (1978) | 214 | 3 | p, n fixed | formula |
| Lanczos3 | NIST StRD (1978) | 24 | 6 | p, n fixed | formula |
| Gauss1 | NIST StRD (1978) | 250 | 8 | p, n fixed | formula |
| Gauss2 | NIST StRD (1978) | 250 | 8 | p, n fixed | formula |
| DanWood | NIST StRD (1978) | 6 | 2 | p, n fixed | formula |
| Misra1b | NIST StRD (1978) | 14 | 2 | p, n fixed | formula |
| Kirby2 | NIST StRD (1978) | 151 | 5 | p, n fixed | formula |
| Hahn1 | NIST StRD (1978) | 236 | 7 | p, n fixed | formula |
| Nelson | NIST StRD (1978) | 128 | 3 | p, n fixed | formula |
| MGH17 | NIST StRD (1978) | 33 | 5 | p, n fixed | formula |
| Lanczos1 | NIST StRD (1978) | 24 | 6 | p, n fixed | formula |
| Lanczos2 | NIST StRD (1978) | 24 | 6 | p, n fixed | formula |
| Gauss3 | NIST StRD (1978) | 250 | 8 | p, n fixed | formula |
| Misra1c | NIST StRD (1978) | 14 | 2 | p, n fixed | formula |
| Misra1d | NIST StRD (1978) | 14 | 2 | p, n fixed | formula |
| Roszman1 | NIST StRD (1978) | 25 | 4 | p, n fixed | formula |
| ENSO | NIST StRD (1978) | 168 | 9 | p, n fixed | formula |
| MGH09 | NIST StRD (1978) | 11 | 4 | p, n fixed | formula |
| Thurber | NIST StRD (1978) | 37 | 7 | p, n fixed | formula |
| BoxBOD | NIST StRD (1978) | 6 | 2 | p, n fixed | formula |
| Ratkowsky2 | NIST StRD (1978) | 9 | 3 | p, n fixed | formula |
| MGH10 | NIST StRD (1978) | 16 | 3 | p, n fixed | formula |
| Eckerle4 | NIST StRD (1978) | 35 | 3 | p, n fixed | formula |
| Ratkowsky3 | NIST StRD (1978) | 15 | 4 | p, n fixed | formula |
| Bennett5 | NIST StRD (1978) | 154 | 3 | p, n fixed | formula |
| Isomerization | Bates and Watts (1988) | 24 | 4 | p, n fixed | formula |
| Lubricant | Bates and Watts (1988) | 53 | 9 | p, n fixed | formula |
| Sulfisoxazole | Bates and Watts (1988) | 12 | 4 | p, n fixed | formula |
| Leaves | Bates and Watts (1988) | 15 | 4 | p, n fixed | formula |
| Chloride | Bates and Watts (1988) | 54 | 3 | p, n fixed | formula |
| Tetracycline | Bates and Watts (1988) | 9 | 4 | p, n fixed | formula |
| Linear, full rank | Moré, Garbow, and Hillstrom (1981) | 10 | 5 | p <= n free | function |
| Linear, rank 1 | Moré, Garbow, and Hillstrom (1981) | 10 | 5 | p <= n free | function |
| Linear, rank 1, zero columns and rows | Moré, Garbow, and Hillstrom (1981) | 10 | 5 | p <= n free | function |
| Rosenbrock | Moré, Garbow, and Hillstrom (1981) | 2 | 2 | p, n fixed | function |
| Helical valley | Moré, Garbow, and Hillstrom (1981) | 3 | 3 | p, n fixed | function |
| Powell singular | Moré, Garbow, and Hillstrom (1981) | 4 | 4 | p, n fixed | function |
| Freudenstein/Roth | Moré, Garbow, and Hillstrom (1981) | 2 | 2 | p, n fixed | function |
| Bard | Moré, Garbow, and Hillstrom (1981) | 15 | 3 | p, n fixed | function |
| Kowalik and Osborne | Moré, Garbow, and Hillstrom (1981) | 11 | 4 | p, n fixed | function |
| Meyer | Moré, Garbow, and Hillstrom (1981) | 16 | 3 | p, n fixed | function |
| Watson | Moré, Garbow, and Hillstrom (1981) | 31 | 6 | p, n fixed | function |
| Box 3-dimensional | Moré, Garbow, and Hillstrom (1981) | 10 | 3 | p <= n free | function |
| Jennrich and Sampson | Moré, Garbow, and Hillstrom (1981) | 10 | 2 | p <= n free | function |
| Brown and Dennis | Moré, Garbow, and Hillstrom (1981) | 20 | 4 | p <= n free | function |
| Chebyquad | Moré, Garbow, and Hillstrom (1981) | 9 | 9 | p <= n free | function |
| Brown almost-linear | Moré, Garbow, and Hillstrom (1981) | 10 | 10 | p == n free | function |
| Osborne 1 | Moré, Garbow, and Hillstrom (1981) | 33 | 5 | p, n fixed | function |
| Osborne 2 | Moré, Garbow, and Hillstrom (1981) | 65 | 11 | p, n fixed | function |
| Hanson 1 | Salane (1987) | 16 | 2 | p, n fixed | function |
| Hanson 2 | Salane (1987) | 16 | 3 | p, n fixed | function |
| McKeown 1 | McKeown (1975) | 3 | 2 | p, n fixed | function |
| McKeown 2 | McKeown (1975) | 4 | 3 | p, n fixed | function |
| McKeown 3 | McKeown (1975) | 10 | 5 | p, n fixed | function |
| Devilliers and Glasser 1 | Salane (1987) | 24 | 4 | p, n fixed | function |
| Devilliers and Glasser 2 | Salane (1987) | 16 | 5 | p, n fixed | function |
| Madsen example | Madsen (1988) | 3 | 2 | p, n fixed | function |
For each test problem, the data, nonlinear model and target parameter values can be retrieved using nls_test_problem(), as also illustrated above for the Gauss1 and Lubricant datasets. The nls_test_problem() function includes suggested starting values for all regression problems and for optimization problems when using the default number of parameters and residuals (p = NA, n = NA). For the optimization problems, a function calculating the \(n \times p\) Jacobian matrix is also returned. This function can be passed to the jac argument of gsl_nls() in order to use analytic evaluation of the gradient in the NLS algorithm.
Example regression problem (Misra1a)
## nls model + data
(misra1a <- nls_test_problem("Misra1a"))
#> $data
#> y x
#> 1 10.07 77.6
#> 2 14.73 114.9
#> 3 17.94 141.1
#> 4 23.93 190.8
#> 5 29.61 239.9
#> 6 35.18 289.0
#> 7 40.02 332.8
#> 8 44.82 378.4
#> 9 50.76 434.8
#> 10 55.05 477.3
#> 11 61.01 536.8
#> 12 66.40 593.1
#> 13 75.47 689.1
#> 14 81.78 760.0
#>
#> $fn
#> y ~ b1 * (1 - exp(-b2 * x))
#> <environment: 0x562f11445798>
#>
#> $start
#> b1 b2
#> 5e+02 1e-04
#>
#> $target
#> b1 b2
#> 2.389421e+02 5.501564e-04
#>
#> attr(,"class")
#> [1] "nls_test_formula"
## nls model fit
with(misra1a,
gsl_nls(
fn = fn,
data = data,
start = start
)
)
#> Nonlinear regression model
#> model: y ~ b1 * (1 - exp(-b2 * x))
#> data: data
#> b1 b2
#> 2.389e+02 5.502e-04
#> residual sum-of-squares: 0.1246
#>
#> Algorithm: multifit/levenberg-marquardt, (scaling: more, solver: qr)
#>
#> Number of iterations to convergence: 16
#> Achieved convergence tolerance: 6.745e-15Example optimization problem (Rosenbrock)
## nls model fit
with(nls_test_problem("Rosenbrock"),
gsl_nls(
fn = fn,
y = y,
start = start,
jac = jac
)
)
#> Nonlinear regression model
#> model: y ~ fn(x)
#> x1 x2
#> 1 1
#> residual sum-of-squares: 9.762e-19
#>
#> Algorithm: multifit/levenberg-marquardt, (scaling: more, solver: qr)
#>
#> Number of iterations to convergence: 16
#> Achieved convergence tolerance: 7.696e-14Benchmark NLS fits
To conclude, we benchmark the performance of the multistart algorithm by computing NLS model fits for each of the 59 test problems using the multistart algorithm with no starting values provided, i.e. all starting values are set to NA. As trust region method we choose respectively: the default Levenberg-Marquardt algorithm (algorithm = "lm"); the double dogleg algorithm (algorithm = "ddogleg"); and the Levenberg-Marquardt algorithm with geodesic acceleration (algorithm = "lmaccel"). The maximum number of allowed iterations maxiter is set to \(10\ 000\), all other tuning parameters in the control argument are kept at their default values according to gsl_nls_control(). For comparison, we also compute single-start NLS model fits using the default Levenberg Marquardt algorithm (algorithm = "lm"), with as naive choice of starting values a vector of all ones \((1, \ldots, 1)\), similar to nls() when argument start is missing.
The table below displays the NLS model fit results for each individual test problem using the following status colors:
- success ; the NLS routine converged successfully and the residual sum-of-squares (SSR) under the fitted parameters coincides (up to a small numeric tolerance) with the SSR under the target parameter values4. The total runtime is displayed in seconds, timed on a modern laptop computer (Intel i7-8550U CPU, 1.80GHz) using a single core.
- false convergence ; the NLS routine converged successfully, but the residual SSR under the fitted parameters is larger than the SSR under the target parameter values.
- max. iterations ; the NLS routine failed to converge within the maximum number of allowed iterations.
- failed/non-zero exit ; the NLS routine failed to converge and returns an error or an NLS object with a non-zero exit code.
We observe that the naive single-start model fits manage to correctly fit about half of the test problems (27 out of 59), suggesting that these test problems are straightforward to optimize and do not require well-informed starting values. The multistart model fits using the double dogleg method improve upon the naive single-start model fits achieving correct convergence for 51 out of 59 test problems. The multistart Levenberg-Marquardt model fits correctly converge for a few more test problems (56 out of 59). Finally, the most robust results are obtained with the multistart model fits using the Levenberg-Marquardt algorithm with geodesic acceleration, which correctly fit all 59 test problems without initializing proper starting values or starting ranges! 🎉
lm/single-start | lm/multi-start | ddogleg/multi-start | lmaccel/multi-start | |
|---|---|---|---|---|
Misra1a | 0.4s | 0.2s | 0.2s | |
Chwirut2 | 0.4s | 0.3s | 0.4s | |
Chwirut1 | 0.5s | 0.4s | 0.7s | |
Lanczos3 | 0.02s | 2s | 2s | 2s |
Gauss1 | 4s | 2s | ||
Gauss2 | 4s | 2s | ||
DanWood | 0.003s | 0.3s | 0.3s | 0.3s |
Misra1b | 0.4s | 0.6s | 0.6s | |
Kirby2 | 0.01s | 0.3s | 0.3s | 0.4s |
Hahn1 | 2s | |||
Nelson | 0.3s | 0.6s | 0.2s | |
MGH17 | 0.9s | 0.3s | 1s | |
Lanczos1 | 0.02s | 3s | 2s | 1s |
Lanczos2 | 0.02s | 2s | 2s | 2s |
Gauss3 | 4s | 2s | ||
Misra1c | 0.8s | |||
Misra1d | 2s | 2s | 0.5s | |
Roszman1 | 4s | 5s | ||
ENSO | 6s | 5s | 5s | |
MGH09 | 0.004s | 0.4s | 0.4s | 0.5s |
Thurber | 1s | 1s | 2s | |
BoxBOD | 0.3s | 0.2s | 0.4s | |
Ratkowsky2 | 0.1s | 0.1s | 0.3s | |
MGH10 | 2s | 0.7s | 1s | |
Eckerle4 | 0.4s | 0.4s | 0.5s | |
Ratkowsky3 | 0.4s | 0.3s | 0.4s | |
Bennett5 | 1s | |||
Isomerization | 0.008s | 0.6s | 0.6s | 0.6s |
Lubricant | 0.01s | 2s | 2s | 3s |
Sulfisoxazole | 0.3s | 0.4s | 0.5s | |
Leaves | 1s | 0.9s | 0.3s | |
Chloride | 0.3s | 0.4s | 0.6s | |
Tetracycline | 0.005s | 0.5s | 0.3s | 0.4s |
Linear, full rank | 0.002s | 0.2s | 0.2s | 0.2s |
Linear, rank 1 | 0.002s | 0.5s | 0.3s | 0.4s |
Linear, rank 1, zero columns and rows | 0.002s | 0.4s | 0.3s | 0.3s |
Rosenbrock | 0.002s | 0.3s | 0.1s | 0.3s |
Helical valley | 0.002s | 0.5s | 0.2s | 0.3s |
Powell singular | 0.002s | 0.3s | 0.2s | 0.3s |
Freudenstein/Roth | 0.4s | 0.2s | 0.4s | |
Bard | 0.001s | 0.3s | 0.2s | 0.3s |
Kowalik and Osborne | 0.002s | 0.3s | 0.2s | 0.2s |
Meyer | 1s | 0.5s | 1s | |
Watson | 0.004s | 0.9s | 0.4s | 0.5s |
Box 3-dimensional | 0.002s | 0.1s | 0.2s | 0.4s |
Jennrich and Sampson | 0.003s | 0.2s | 0.2s | 0.3s |
Brown and Dennis | 0.008s | 0.6s | 0.5s | 0.7s |
Chebyquad | 0.009s | 1s | 0.5s | 1s |
Brown almost-linear | 0.001s | 0.7s | 0.6s | 0.9s |
Osborne 1 | 0.4s | 0.2s | 0.5s | |
Osborne 2 | 1s | 1s | ||
Hanson 1 | 0.2s | 0.2s | 0.4s | |
Hanson 2 | 0.2s | 0.2s | 0.2s | |
McKeown 1 | 0.002s | 0.2s | 0.2s | 0.3s |
McKeown 2 | 0.003s | 0.3s | 0.2s | 0.4s |
McKeown 3 | 0.004s | 0.4s | 0.3s | 0.4s |
Devilliers and Glasser 1 | 0.4s | 0.4s | 0.6s | |
Devilliers and Glasser 2 | 0.6s | 0.7s | 1s | |
Madsen example | 0.004s | 0.2s | 0.2s | 0.3s |
# Successful fits | 27/59 | 56/59 | 51/59 | 59/59 |
References
a basin of attraction refers to the basin surrounding a local minimum of the objective function, such that starting from any point inside the basin the local optimizer converges to the same local minimum.↩︎
\(f^+ = f \vee 0\) and \(f^- =-f \vee 0\) denote the positive and negative part of \(f\).↩︎
The reason for using the residual sum-of-squares (SSR) to check for correct convergence of the NLS model fits is that several problems have multiple distinct parameter solutions that result in the same (optimal) SSR.↩︎